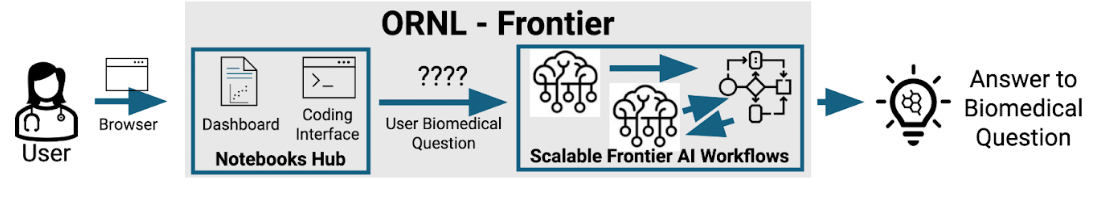
Using the latest in open Large Language Models (LLMs) to power biomedical search, discovery, and understanding from the literature securely and at scale.
The overarching goal of this project is to enable researchers to search and synthesize all biomedical information their institution subscribes to along with publicly available data using the latest in open Large Language Model (LLM) technologies. To enable this, the Oak Ridge Leadership Computing Facility, home of ORNL’s Frontier Exascale supercomputer, will host the “Ask AIthena” application. “Ask AIthena” utilizes state-of-the-art open LLMs to transform biomedical journal articles, pre-print articles, textbooks, clinical databases, etc. into a format that is easy to search and understand and allows users to ask questions from that large text corpus. This approach ensures that copywritten material is never shared or used to train a model and that users are only enabled to search and synthesize the subset of content that their institution subscribes to. “Ask AIthena” provides a simple user interface for non-technical users to ask questions, find relevant citations to their query, and have in-depth discussions about the relevant text corpus to understand a topic thoroughly.
The pilot program will only process publicly available abstracts from PubMed and all full-text articles from Arxiv, Chemrxiv, Biorxiv, and Medrxiv. However, this limitation is purely to demonstrate that the approach is feasible at scale and that different sources of information and publishers can be subset easily per user such that only relevant and provisioned information is returned to users.