The ability to design novel proteins tailored for specific purposes will transform humankind. Currently, improving AI models using real-world experimental data is still a challenge. Key hurdles include optimizing AI architectures, efficient training based on validated data, and incorporating diverse data sources. In addition, computational challenges still exist, leading to performance bottlenecks. One way to overcome these challenges is by combining AI with high-performance computing (HPC), which can significantly accelerate progress scientifically and computationally. The IMPRESS initiative aims to address these challenges by integrating AI and HPC systems to enable real-time model evaluation and more efficient generation of high-quality proteins, driving advancements in protein design.
IMPRESS: Integrated Machine-learning for PRotein Structures at Scale (EAGER)
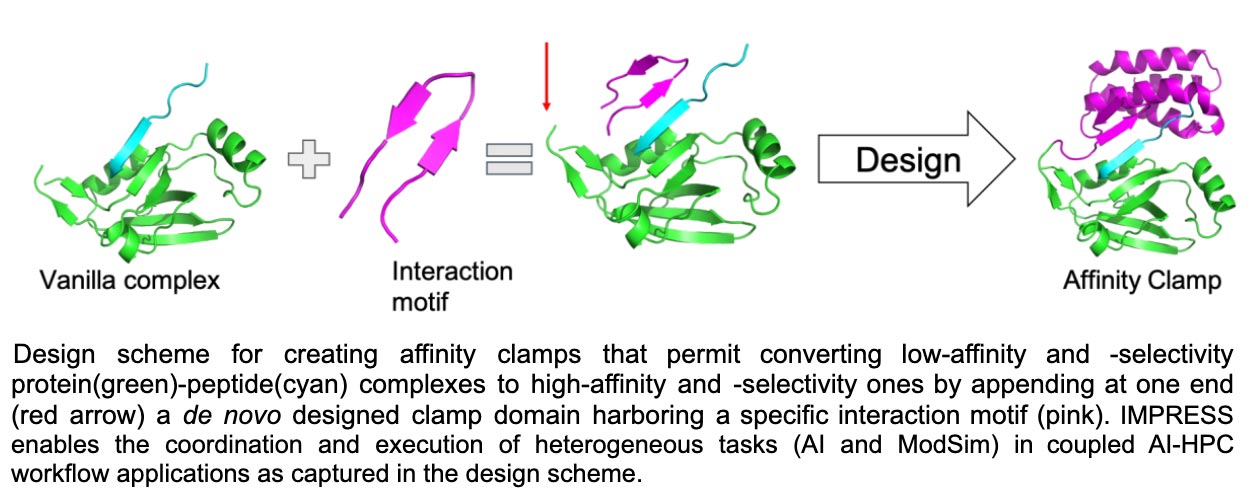
Project leads, key team members
Shantenu Jha (Principal Investigator, Rutgers University), Sagar Khar (Principal Investigator, Rutgers University), Matteo Turilli (Co-Principle Investigator, Rutgers University), Mikhail Titov (Assistant Computational Scientist, Brookhaven National Lab (BNL)), Aymen Alsaadi (Research associate, Rutgers University), Andre Merzky (Senior Research Programmer, Rutgers University), Jonathan Ash (PhD Student, Rutgers University), Ozgur Ozan Kilic (Research Scientist, Brookhaven National Lab (BNL))
Advancing NAIRR Infrastructure
The project focuses on integrating AI and HPC systems to support the online coupling of AI-HPC on the NAIRR platform. The architecture and infrastructure will extend NAIRR pilot resources without restricting them to traditional HPC platforms. The project aims to build novel capabilities on NAIRR, creating a scalable, concurrent AI-HPC infrastructure for real-time simulations and model optimization.
Advancing AI research
The project aims to enhance AI's role in protein design by combining AI-driven generative models with HPC simulations. This coupling will allow real-time feedback between AI and HPC tasks, improving the design and generation of proteins and foundational models through experimental data validation. The project aims to achieve 100x and 1000x acceleration over existing "vanilla" approaches.
Broader Impacts
IMPRESS project has three main broader impacts: education, capacity building, and broadening participation in STEM. The project will integrate developed prototypes and software systems into curricula, offering students and researchers, including non-computing individuals, exposure to advanced AI and HPC tools. A specific course, "AI-enabled protein science and engineering," will be provided for graduate Bioscience students. In collaboration with the RCSB Protein DataBank, the project will develop an "Introduction to Protein Design using HPC" crash course, primarily targeting underrepresented minority undergraduate students through the Rutgers Research-Intensive Summer Experience (RISE) program. Developing the crash course will also be extended to New Jersey's primarily undergraduate institutions (PUIs), further promoting diversity and inclusion in STEM fields.
More information
Learn about how IMPRESS: Integrated Machine-learning for PRotein Structures at Scale can meet your needs by contacting our team directly:
- shantenu.jha@rutgers.edu or khare@chem.rutgers.edu. Visit our website to learn more at: https://radical-project.github.io/impress/.
This work is supported by National Science Foundation Grant No. (#2438557).